Reproductive Microbiomes: Using the Microbiome as a Novel Diagnostic Tool for Endometriosis
Melissa A. Cregger, Katherine Lenz, Elizabeth Leary, Richard Leach, Asgerally Fazleabas, Bryan White, Andrea Braundmeier
DOI10.21767/2476-1974.100036
Melissa A. Cregger1,2, Katherine Lenz3, Elizabeth Leary5,6, Richard Leach5,6, Asgerally Fazleabas5,6, Bryan White1, Andrea Braundmeier1,3,4*
1Carl R Woese Institute for Genomic Biology, The University of Illinois, 1206 W. Gregory Dr. Urbana, IL 61801, USA
2Biosciences Division, Oak Ridge National Laboratory, 1 Bethel Valley RD, Oak Ridge TN 37831, USA
3Department of Medical Microbiology, Immunology and Cell Biology, Southern Illinois University School of Medicine, Springfield, IL 62702, USA
4Department of Obstetrics and Gynecology, Southern Illinois University School of Medicine, Springfield, IL 62702, USA
5Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State College of Medicine, Grand Rapids, MI, USA
6Spectrum Health Medical Group, Grand Rapids, MI, USA
- *Corresponding Author:
- Andrea Braundmeier
Department of Obstetrics and Gynecology
Southern Illinois University School of Medicine, Springfield, IL 62702, USA
Tel: +217-545-5226
E-mail: abraundmeier88@siumed.edu
Received date: September 4, 2017; Accepted date: September 21, 2017; Published date: September 25, 2017
Citation: Cregger MA, Braundmeier A, Lenz K, Leary E, Leach R, et al. (2017) Reproductive Microbiomes: Using the Microbiome as a Novel Diagnostic Tool for Endometriosis. Reproduct Immunol Open Access Vol.2 Iss.3:36.doi: 10.21767/2476-1974.100036
Copyright: © 2017 Cregger MA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Endometriosis is a chronic inflammatory disease which results in significant pain and long term reproductive consequences for up to 50% of infertile women. This study was focused to understand how endometriosis altered the uterine and cervical bacterial community. Methods and findings: Urogenital swabs and uterine washes were collected from 19 pre-menopausal women undergoing laparoscopic surgery for pelvic pain, suspected endometriosis (experimental n=10), and women undergoing laparoscopic surgery for benign ovarian/uterine conditions (control n=9). Patients were followed for the next year and repeat cervical swabs were obtained. Bacterial community composition was assessed from these samples using Illumina next generation 16S rRNA amplicon sequencing. Bacterial communities were significantly different between sample sites, the uterus and cervix, and stage III endometriosis resulted in significant alterations in the cervical bacterial community. Both bacterial richness and phylogenetic diversity increased in association with stage III endometriosis. Surgical intervention resulted in a stabilized cervical bacterial community for a short period of time. Conclusion: Bacterial community profiling may provide a useful diagnostic tool for identifying endometriosis in asymptomatic, infertile women in a clinical setting.
Keywords
Cervix; endometriosis; Microbiome; Next generation sequencing; Uterus
Abbreviations
OUT: Operational Taxonomic Unit; PCR: Polymerase Chain Reaction
Introduction
In recent years, it has become evident that the archaea, bacteria, and fungi (collectively the microbiome) that inhabit the human body play key roles in regulating a variety of functions like digestion, immune responses [1], resistance to cancers [2], as well as modulating environmental conditions within the human body like pH [3]. Perturbations to the human microbiome can result in a myriad of disease states and conversely, a variety of diseases or factors can alter the microbiota inhabiting a given site. Understanding complex interactions between the human host and the microbiome has become incredibly important to predict and understand a variety of diseases. Organs within the human body once believed to be sterile have now been shown to host a diverse assemblage of microorganisms. This is apparent within the female reproductive tract [4]. The uterus was once believed to be a sterile environment, but it has recently been shown that numerous, phylogentically diverse organisms may inhabit this organ [5-7]. Understanding fluctuations in these organisms may be important for a variety of disease states that influence fertility like polycystic ovarian syndrome, endometriosis, and a variety of bacterial infections.
Endometriosis is a chronic inflammatory gynaecological disease characterized by the presence of endometrial tissue in ectopic locations and affecting up to 10% of reproductive-aged women and up to 50% of women who experience infertility [8,9]. The current method for diagnosing endometriosis requires invasive laparoscopic surgery contributing to delayed diagnosis by approximately 10 years from the onset of disease, allowing significant progression before women begin treatment potentially resulting in decreased fertility. Endometriotic lesions have the ability to develop on the peritoneal surface of pelvic organs and can cause the development of severe adhesions within the pelvic cavity resulting in pain and also organ dysfunction. Historically, the staging of endometriosis has been calculated by the location of lesion development in the peritoneal cavity, severity of pelvic adhesions, and involvement of the ovaries [10]. However, the rASRM staging criteria is limited by visual detection at the time of surgery and does not take into consideration pelvic organ involvement and deeply infiltrating endometriosis. Therefore, the ENZIAN staging criteria was developed that incorporated disease involvement of peritoneal structures and DIE to be used as a supplement to the rASRM [11]. Neither of these staging criteria account for patient factors such as pain and infertility and therefore cannot be used to estimate patient fertility status. Therefore, the Endometriosis Fertility Index (EFI) was developed for clinicians to predict patient fertility status in those patients with surgically documented disease [12]. While the development of these staging models benefits clinical classification of individual patient disease, they do not provide information on how disease pathogenesis and how disease development at each staging level affects other physiological systems. Additionally, it is unclear how the formation of these lesions alters the microbiota inhabiting the peritoneal cavity, uterus, cervix, or vagina.
It is well established that the peritoneal cavity of women with endometriosis is an inflammatory environment. Patients with endometriosis have elevated levels of immune mediators in the peritoneal fluid, which is believed to occur as a result of improper ectopic tissue clearance [13-15]. Women with endometriosis have been shown to have reduced T regulatory cell populations in both the periphery and within the endometrium [16], as well as increased T regulatory cells in ectopic endometrial tissue [17], creating an immunosuppressive microenvironment that allows for endometriotic lesions to survive. Together, these data suggest endometriosis may be characterized as an immune disorder with autoimmune and chronic inflammatory tendencies. Aberrant immune responses in the endometrium and peritoneal cavity may facilitate decreased fertility; 30-50% of women with endometriosis suffer from infertility due to various reproductive irregularities throughout oocyte maturation and implantation [18,19]. Much of this immune dysfunction in the reproductive tract can be attributed to inflammation caused by the presence of ectopic lesions.
Across the human body, inflammatory disease have been linked to alterations in the microbiome [1]. This may be due to the presence of one pathogenic organism like Helicobacter pylori that is known to cause stomach ulcers, gastritis, and gastric cancer [20] and potentially increase the likelihood of autoimmune diabetes [21], or may be due to microbial community level shifts. Within the gut, it has been demonstrated that inflammatory bowel disease may result from altered interactions between the intestinal microbes and the mucosal immune system [22]. It is hypothesized that a variety of inflammatory diseases with unknown etiology, like rheumatoid arthritis, may be a result of microbes entering the blood from the gut or oral cavity and causing disease [23]. Further, early perturbations of the gut microbiota may result in chronic inflammation and metabolic disease [24]. Thus far, very few studies have been conducted to identify how inflammation in the female reproductive tract may alter the microbiome.
Immune dysfunction and chronic inflammation in women with endometriosis may have direct impacts on the microorganisms associated with organs in the reproductive tract, specifically the uterus and the cervix. Recent research has demonstrated that there are robust and diverse uterine [5] and cervical [25] bacterial communities. Studies are now beginning to examine how different diseases may cause deviations in these communities. Indicator taxa are being identified that may help clinicians diagnose enigmatic diseases like endometrial cancer [2], but thus far this has remained elusive for endometriosis. Using next generation amplicon sequencing of the bacterial 16S rRNA gene, this pilot study is poised to identify how endometriosis alters the uterine and cervical bacterial community. Further, this study aims to identify indicator taxa within the cervix that may help clinicians diagnose active endometriosis through the use of a cervical swab, avoiding unnecessary laparoscopic surgery as a means of diagnosis. We hypothesized that patients with endometriosis would have altered uterine and cervical bacterial communities and the most pronounced effect would be seen in patients with advanced endometriosis.
Methods
Subjects
The patient recruitment and sample collection protocols were approved by the Institutional Review Boards (IRBs) of Michigan State University (Protocol#: 07-712) and Southern Illinois University School of Medicine (Protocol#: 14-146). A total of 19 patients were recruited from the Department of Obstetrics and Gynecology at Spectrum Health Medical Group (EL) and referred to the study coordinator upon showing interest in this pilot study. Pre-menopausal women undergoing laparoscopic surgery for pelvic pain with suspicion or known endometriosis were enrolled for the experimental group (n=10; stage I: 6, stage II: 1, stage III: 1, stage IV: 2), while women scheduled for a laparoscopy/laparotomy/hysterectomy for benign uterine or ovarian conditions were enrolled as controls (n=9). Women in the control group were examined during surgery to rule out the presence of endometriotic lesions. Cervical swabs and uterine washes were acquired while the patient was under anesthesia. For the experimental group, confirmation and staging of endometriosis (stages I-IV) was completed using the rASRM classification scale. Stage of menstrual cycle was recorded by last menstrual period. Exclusion criteria included, current or previous hormone treatment within the last 3 months, IUD within the previous 3 months, presence of pelvic inflammatory disease at laparoscopy, and women who no longer have menstrual cycles.
Sample collection
All sample collection was performed by standard operating procedures and followed the protocol established by the approved IRB and Human Microbiome Project (HMP) [26]. A cervical swab and a uterine wash was taken on the day of surgery for each patient in both treatment groups. Cervical swabs were then taken for each patient at two weeks postsurgery, 3-4 months post-surgery, and 10-14 months postsurgery (Table 1). All samples were frozen at -80° C until analysis.
DNA extraction
DNA was extracted from the cervical swabs and uterine washes using the Power Soil® DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer’s instructions. After extraction, the DNA stock concentration was measured using a NanoDrop-2000 spectrophotometer (Thermo Scientific, Inc., Wilmington, DE), with samples ranging from 1.03 to 22.4 ng/μl.
16S rRNA gene amplification and sequencing
A 584 bp fragment of the hypervariable V3-V5 region of the 16S rRNA gene was amplified by a polymerase chain reaction (PCR) as follows: 25 μl Kapa HiFi (Kapa Biosystems, Woburn, MA), 25 μM forward primer, 25 μM reverse primer, 50 ng of DNA, and molecular grade water to reach a final volume of 50 μl per reaction.
The 357F forward primer was used read 1: 5’TATGGTAATTGTCCTACGGGAGGCAGCAG3’;
read 2: 5’AGTCAGTCAGCCCCGTCAATTCMTTTRAGT3’;
index: 5’ACTYAAAKGAATTGACGGGGCTGACTGACT3’.
The universal reverse primer was 926R (5’CCGTCAATTCMTTTRAGT3’). One of 96 specific barcodes consisting of 12 base pairs was added to the reverse primer for each sample. The PCR cycle was as follows: 45 seconds at 98° C followed by 25 cycles of 15 seconds at 98° C, 30 seconds at 65° C, and 30 seconds at 72° C for denaturation, and then 2 minutes at 72° C and hold at 4° C for final extension, as described in the HMP [26]. The PCR products were then run on an agarose gel, where it was determined that additional PCR amplification utilizing the same initial primer set was required. Following PCR amplification, the products were then purified using the Gene JET PCR Purification Kit (Thermo Scientific, Inc., Wilmington, DE). Final DNA product concentrations were measured by the NanoDrop-2000 spectrophotometer and were between 41.2 and 132.2 ng/μl. Samples were then pooled together by mass (15 μg), with one pool consisting of samples containing reverse primer barcodes 1-96. Total pool concentration was measured by Qubit 2.0 Fluorometer (Invitrogen, Life Technologies, Carlsbad, CA). All pools were then sent to the University of Illinois at Urbana-Champaign to be sequenced utilizing a highthroughput platform (MiSeq; Illumina Inc., San Diego, CA). Approximately 14,000,000 total sequence reads were obtained.
Sequence analysis
Sequences were processed using a custom multiple alignment tool known as the Illinois-Mayo Taxon Operations for RNA Dataset Organization (IM-TORNADO, [27]). This tool merged paired end reads into a single alignment and clustered OTUs (operational taxonomic units) using Abundant OTU+ [28] at 97% sequence similarity.
Sequencing statistics
A total of 4,560,131 sequences were obtained after quality filtering and sequence processing. Across samples, the minimum number of sequences obtained was 0 and the maximum number obtained was 895,925. The average number of sequences per sample was 101,336. Data visualization and summary was performed using QIIME [29]. The raw sequence data are available in the Sequence Read Archive at the National Center for Biotechnology Information (BioProject ID: PRJNA387551).
Data analysis
Before any statistical tests were completed, the dataset was rarefied to 400 sequences per sample to account for variation in sequencing depth. This resulted in the removal of 2 uterine samples and 2 cervical samples from the dataset. Furthermore, one patient’s (H1331) cervical sample was removed from the dataset as an outlier because her cervical bacterial community was 81% Sneathia sp. indicative of bacterial vaginosis [30]. At 400 sequences per sample, rarefaction curves plateaued demonstrating that sufficient sequencing was conducted to fully characterize the bacterial community. Alpha-diversity and Faith’s phylogenetic diversity were calculated for all samples and betadiversity was assessed using both weighted and unweighted UNIFRAC values in QIIME. Weighted UNIFRAC incorporates both taxa identities and relative abundances of organisms while unweighted UNIFRAC simply incorporates differences in taxa identities. Permutational MANOVA tests were performed to understand if bacterial communities differed by organ (cervix vs uterus), between patients, disease stage, and through time. Uterine and cervical bacterial communities were indeed significantly different, so uterine and cervical data were divided and subsequent analyses were performed to identify the effect of disease and time on bacterial community composition. Kruskal Wallis tests were performed to identify OTUs that varied significantly between organs and disease stages.
Results
A total of 18 patients were included in this study once samples were removed as described above due to insufficient sequence numbers or due to the presence of patient infection. 9 patients had uterine samples on the day of surgery and only seven of these samples could be used for subsequent analyses. Two of the samples had insufficient sequence numbers. 17 cervical samples were included in the analysis for the day of surgery after one sample was removed due to insufficient sequence numbers. Repeat cervical sampling varied by patient (Table 1).
| Patient ID | Disease Stages | UT Sample | CVX Swab | CVX Swab | CVX Swab | CVX Swab | CVX Swab |
|---|---|---|---|---|---|---|---|
| H1269 | - | DOS | DOS | ||||
| H1331 | - | DOS | |||||
| H1337 | - | DOS | DOS | ||||
| H1357 | - | DOS | |||||
| H1360 | - | DOS | |||||
| H1365 | - | DOS | |||||
| H1379 | - | DOS | 2 weeks | 4 months | |||
| H1383 | - | DOS | DOS | ||||
| H1409 | - | DOS | 2 weeks | ||||
| H1274 | I | DOS | 10 days | 4 months | 14 months | ||
| H1291 | I | DOS | 2 weeks | ||||
| H1361 | I | DOS | |||||
| H1385 | I | DOS | 2 weeks | ||||
| H1392 | I | DOS | DOS | 2 weeks | |||
| H1402 | I | DOS | DOS | 2 weeks | |||
| H1323 | II | DOS | DOS | 3 weeks | 5 months | ||
| H1272 | III | DOS | DOS | 2 weeks | 4 months | 10 months | 15 months |
| H1223 | IV | DOS | DOS | 2 weeks | |||
| H1334 | IV | DOS | DOS |
Table 1: Patient cervical and uterine sampling scheme and disease stage.
On the day of surgery, uterine and cervical samples were significantly different from one another in terms of both the taxa present and the relative abundance of those taxa (Figure 1; weighted unifrac; pseudo F=4.97, p=0.031; unweighted unifrac pseudo F=2.50, 0.052). Across all samples, there are 174 OTUs found in the uterus and 163 OTUs found in the cervix. 98 of these OTUs overlapped the two organs. Individual uterine samples, on average have 50 OTUs (+/-23.99 std error), while cervical samples have only 20 OTUs (+/-22.60 std error; t=2.72, p=0.011). Not surprisingly, Faith’s phylogenetic diversity followed a similar pattern. Uterine bacterial communities were more phyogenetically diverse (FPD=7.27+/-3.21) than cervical bacterial communities (3.26+/-2.72; t=2.97, p=0.006). The cervical samples were dominated by Firmicutes (93%) and Bacteroidetes (7%). The uterus was 47% Firmicutes, 43% Bacteroidetes, and 10% Proteobacteria. On the day of surgery, the taxa present in the uterus were not significantly different across disease stages (unweighted unifrac; pseudo F=1.97, p=0.216, weighted unifrac; pseudo F=0.74, p=0.571). Similarly, the cervical communities were not different across disease stages on the day of surgery (unweighted unifrac; pseudo F=0.982, p = 0.498, weighted unifrac; pseudo F=1.353, p=0.282).
Patient IDs are followed by A to represent the cervix on the day of surgery or UA to represent the uterus on the day of surgery. Genera are shown that have greater than 5 representative sequences (>1.25% relative abundance) across samples with yellow indicating low abundance and darker colors indicating higher abundance. Absolute abundance is shown after the dataset was rarefied to 400 sequences per sample.
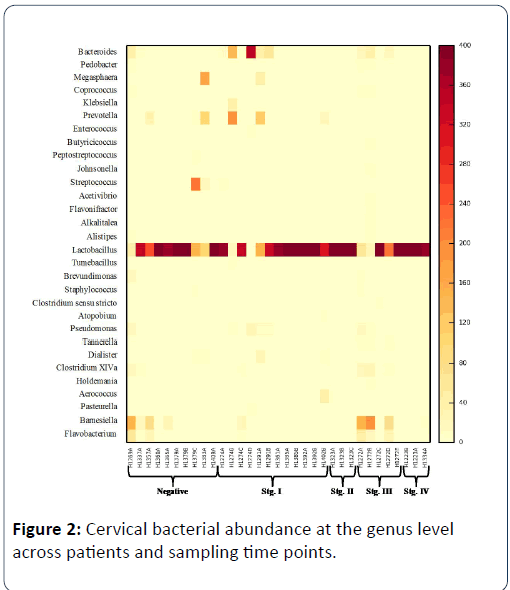
When examining cervical communities through time, there are significant differences in the abundance of bacterial taxa between patients (weighted unifrac; pseudo F=1.16, p=0.372; unweighted unifrac, pseudo F=2.37, p=0.001) but no consistent pattern in bacterial community composition for all patients through time (weighted unifrac; pseudo F=1.33, p=0.233; unweighted unifrac; pseudo F=0.789, p=0.745). Across disease stages, the taxa present varied significantly (weighted unifrac; pseudo F=1.34, p=0.231, unweighted unif rac; pseudo F=1.77, p=0.041), but there was no significant interaction between disease stage and time (weighted unifrac; pseudo F=1.12, p=0.375, unweighted unifrac; pseudo F=0.976, p=0.517). The stage III patient exhibited a very different bacterial community on the DOS relative to all of the other patients. When separating the stage III patient from all other patients, there is a significant effect of disease stage (stage III) and time on bacterial community composition (weighted unifrac; pseudo F=2.49, p=0.053; unweighted unifrac; pseudo F=1.18, p=0.153) showing that at various time points, stage III patient’s cervical bacterial communities are quite different from all other patients Figure 2.
Patient IDs are followed by letters A-E to represent follow up sampling. Genera are shown that have greater than 5 representative sequences (>1.25% relative abundance) across samples with yellow indicating low abundance and darker colours indicating higher abundance. Absolute abundance is shown after the dataset was rarefied to 400 sequences per sample.
On the day of surgery, OTU richness and phylogenetic diversity were greater in the stage III patient relative to all other patients (richness; t=3.48, p=0.016; FPD; t=3.07, p=0.052). Both richness (number of OTUs present) and Faith’s phylogenetic diversity fluctuated through time for the stage III patient. On the day of surgery, there were 58 OTUs (FPD=9.48) detected in the uterus and 79 (FPD=9.9) in the cervix. Two weeks post-surgery, there were 87 OTUs (FPD=10.74) present in the cervix. Four months post-surgery, there were only 5 OTUs (FPD=1.15) detectable in the cervix. Ten months post-surgery, richness increased to 65 OTUs (8.84) present in the cervix, and 15 months post-surgery, richness was down to three OTUs (FPD=0.93). On the day of surgery, there are 56 OTUs that are significantly different between the stage III patient and all other patients (Table 2). Twenty two of these OTUs were only present in the cervix of the stage III patient.
| OTU # | Taxonomy | p | FDR Q | Stage III relative abundance | Other relative abundance |
|---|---|---|---|---|---|
| 92 | Barnesiella | 0.014 | 0.042 | 5.5 | 0.16 |
| 165 | Barnesiella | 0.014 | 0.042 | 4.25 | 0.22 |
| 21 | Barnesiella | 0.004 | 0.019 | 1.75 | 0.03 |
| 154 | Bacteroides | 0.004 | 0.019 | 1.75 | 0.03 |
| 144 | Barnesiella | 0.014 | 0.042 | 1.75 | 0.13 |
| 266 | Staphylococcus | 0.004 | 0.019 | 1.5 | 0.02 |
| 39 | Barnesiella | 0.014 | 0.042 | 1.5 | 0.08 |
| 65 | Barnesiella | 0.014 | 0.042 | 1.5 | 0.09 |
| 129 | Propionibacterium | <0.001 | <0.001 | 1.25 | 0 |
| 35 | Parabacteroides | 0.004 | 0.019 | 1.25 | 0.02 |
| 348 | Coprococcus | 0.004 | 0.019 | 1.25 | 0.03 |
| 363 | Butyricicoccus | <0.001 | <0.001 | 1 | 0 |
| 56 | Barnesiella | 0.004 | 0.019 | 1 | 0.05 |
| 138 | Barnesiella | 0.004 | 0.019 | 1 | 0.03 |
| 232 | Allobaculum | 0.004 | 0.019 | 1 | 0.03 |
| 72 | Tannerella | 0.006 | 0.025 | 1 | 0.06 |
| 88 | Barnesiella | 0.011 | 0.038 | 1 | 0.14 |
| 152 | Clostridium XIVa | 0.014 | 0.042 | 1 | 0.05 |
| 182 | Anaerotruncus | <0.001 | <0.001 | 0.75 | 0 |
| 264 | Acetivibrio | <0.001 | <0.001 | 0.75 | 0 |
| 158 | Achromobacter | 0.006 | 0.025 | 0.75 | 0.05 |
| 79 | Turicibacter | 0.014 | 0.042 | 0.75 | 0.03 |
| 273 | Tannerella | 0.014 | 0.042 | 0.75 | 0.03 |
| 323 | Alkalitalea | 0.014 | 0.042 | 0.75 | 0.05 |
| 282 | Barnesiella | <0.001 | <0.001 | 0.5 | 0 |
| 331 | Lactobacillus | <0.001 | <0.001 | 0.5 | 0 |
| 636 | Coprobacillus | <0.001 | <0.001 | 0.5 | 0 |
| 347 | Ruminococcus | 0.004 | 0.019 | 0.5 | 0.02 |
| 241 | Clostridium XIVa | 0.006 | 0.025 | 0.5 | 0.03 |
| 6 | Sneathia | <0.001 | <0.001 | 0.25 | 0 |
| 213 | Clostridium XIVb | <0.001 | <0.001 | 0.25 | 0 |
| 226 | Ruminococcus | <0.001 | <0.001 | 0.25 | 0 |
| 268 | Clostridium XIVa | <0.001 | <0.001 | 0.25 | 0 |
| 270 | Clostridium XIVa | <0.001 | <0.001 | 0.25 | 0 |
| 283 | Flavonifractor | <0.001 | <0.001 | 0.25 | 0 |
| 311 | Clostridium XIVa | <0.001 | <0.001 | 0.25 | 0 |
Table 2: Taxa present in the cervix of the patient with stage III endometriosis on the day of surgery that demonstrate a significant community level shift associated with disease. A false discovery rate (FDR) correction was used to control for multiple comparisons.
We did not find any effect of stage of the menstrual cycle on the cervical or uterine bacterial communities.
Discussion
The role of the microbiome in the uterus and cervix is largely unknown, but the implications for Human health, reproduction, fertility, and the development of human fetuses are immense [31]. Consistent with other studies characterizing the uterine bacterial community using next generation sequencing, we demonstrated that the uterine bacterial community is comprised mostly of a Bacteroidetes core, with a large percentage of Firmicutes, specifically Lactobacillus sp. that are commonly found in the vagina, although the uterus is not simply a subset of vaginal microbiota [5-7]. Across studies, there is significant variability in the rare taxa detected within the uterine cavity. This variability may be due to timing of sample collection, alterations in hormones which has been shown to change bacterial communities, or dysbiosis in these patient populations due to a variety of disease factors. In our study, approximately half of the patients have endometriosis, and while the control patients do not, they are under a clinician’s care for a variety of symptoms that require laparoscopic surgery, thus these results may not be indicative of a normal uterine bacterial community. Consistent with other studies characterizing the cervical and vaginal bacterial communities, the cervical bacterial communities of our patients were predominately Lactobacillus sp. [25]. Unfortunately, the patients cannot be grouped into traditional vaginal community types [3] because taxonomic resolution at the species level is not reliable, thus genus level characterizations were used. Across samples, numerous low abundance taxa were detected that may be indicative of endometriosis and require further examination.
Classification of stage III (moderate) disease by rASRM criteria indicates that there are significant deep endometriotic lesions on the peritoneum and also ovary. Stage III also indicates the presence of filmy and dense adhesions causing anatomical distortion of the fallopian tubes and ovary, as well as other abdominal structures such as the bowel and bladder [32]. Thus, transition from stage II (mild, superficial lesions) to stage III indicates a significant disease progression which may result from a highly inflammatory abdominal environment. Transition from Stage III to stage IV (Severe) is characterized by a “frozen” abdominal cavity, resultant from the formation of adhesions and inflammatory environment. Essentially it can be thought of as the result of inflammation rather than an active inflammatory process. This theory could explain why the stage III patient had a significantly different bacterial community than all of the other patients. The cervix of this patient harbored a highly diverse bacterial community depleted in the typically predominant Lactobacillus sp. found in the vagina and had increased levels of Firmicutes and Bacteroidetes. We identified 56 OTUs that are significantly different on the day of surgery between the stage III patient and all other patients. Twenty-two of these OTUs are only evident on the cervix of the stage III patient. This community level change may be indicative of severe, active endometriosis and provide further explanation for decreased rates of pregnancy in these women. Indeed, it has been shown that within the vagina, this shift in microorganisms has been linked to decreases in pregnancy as a result of IVF [33], thus a more thorough, detailed characterization of these communities through time is warranted. Examining the cervical bacterial community of the stage III patient upon follow up visits, we showed that the bacterial community was similar to the DOS two weeks post-surgery, but at the four month post-surgery visit, the cervical bacterial community was 97% Lactobacillus sp. with decreased richness and diversity. By ten months post-surgery, the bacterial community was once again highly diverse. This fluctuation in the bacterial community may help explain and predict pregnancy success post-surgery.
In this study, we only characterized bacterial community dynamics in association with endometriosis. It is becoming apparent that other key members of the microbiome like fungi and viruses may play a key role in human health and disease [31] and understanding these dynamics will be imperative if we are to improve these conditions. Future studies will characterize other members of the microbiome to understand how endometriosis alters these communities. Further, future studies will take advantage of the induced non-human primate animal model of endometriosis allowing for the measurement of microbial changes throughout disease progression. This animal model closely resembles lesion kinetics as seen in human disease [34] and use of non-human primates allows for repetitive surgical sampling for multiple time point analysis. Additionally, the inflammatory profile of endometriosis in this animal model mirrors what has been reported in human disease [35-37] making this an excellent parallel study for our currently reported data.
Conclusion
Bacterial community profiling may provide a useful diagnostic tool for identifying endometriosis in asymptomatic, infertile women in a clinical setting.
Source of Funding
This study has been supported by institutional research funds from ABF, MAC, BW, and AF. MAC was supported by a Carl R. Woese Institute for Genomic Biology Postdoctoral Fellowship. MAC is currently supported by a Liane Russell Fellowship at Oak Ridge National Laboratory sponsored by the Laboratory Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle, LLC, for the U.S. Department of Energy.
Acknowledgements
The authors would like to thank two anonymous reviewers for comments on a previous version of this manuscript.
References
- Blaser MJ (2014) The microbiome revolution. Journal of Clinical Investigation 124: 4162-4165.
- Walther Antonio MR, Chen J, Multinu F, Hokenstad A, Distad TJ, et al. (2016) Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Medicine 8: 122.
- Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, et al. (2011) Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America 108: 4680-4687.
- Braundmeier AG, Lenz KM, Chia N, Jeraldo P, Yang F, et al. (2015) Individualized medicine and the microbiome in reproductive tract. Frontiers in Physiology 6.
- Verstraelen H, Vilchez Vargas R, Desimpel F, Jauregui R, Vankeirsbilck N, et al. (2016) Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1-2 region of the 16S rRNA gene. Peerj 4: e1602.
- Franasiak JM, Werner MD, Juneau CR, Tao X, Landis J, et al. (2016) Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet 33: 129-36.
- Moreno I, Codoñer FM, Vilella F, Valbuena D, Martinez Blanch JF, et al. (2016) Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol 215: 684-703.
- Sayasneh AD, Tsivos, Crawford R (2011) Endometriosis and ovarian cancer: a systematic review. ISRN Obstet Gynecol 2011: 140-310.
- D'Hooghe TM, Debrock S, Hill JA, et al. (2003) Endometriosis and subfertility: is the relationship resolved? Semin Reprod Med 21: 243-254.
- American Society for Reproductive Medicine (1997) Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertility and Sterility 67: 817-821.
- Haas D, Shebl O, Shamiyeh A, Schimetta W, Mayer R, et al. (2013) Enzian classification: does it correlate with clinical symptoms and the rASRM score? Acta Obstet Gynecol Scand 92: 562-600.
- Adamson GD (2013) Endometriosis Fertility Index: is it better than the present staging systems? Curr Opin Obstet Gynecol 25: 186-192.
- Hou Z, Sun L, Gao L, Liao L, Mao Y, et al. (2009) Cytokine array analysis of peritoneal fluid between women with endometriosis of different stages and those without endometriosis. Biomarkers 14: 604-618.
- Kyama CM, Mihalyi A, Simsa P, Falconer H, Fulop V, et al. (2009) Role of cytokines in the endometrial-peritoneal cross-talk and development of endometriosis. Front Biosci (Elite Ed) 1: 444-454.
- Tran LV, Tokushige N, Berbic M, Markham R, Fraser IS, et al. (2009) Macrophages and nerve fibres in peritoneal endometriosis. Hum Reprod 24: 835-841.
- Braundmeier A, Jackson K, Koehler J, Nowak R, Fazleabas A, et al. (2012) Induction of endometriosis alters the peripheral and endometrial regulatory T cell population in the non-human primate. Hum Reprod 27: 1712-1722.
- Berbic M, Hey Cunningham AJ, Ng C, Tokushige N, Ganewatta S, et al. (2010) The role of Foxp3+ regulatory T-cells in endometriosis: a potential controlling mechanism for a complex, chronic immunological condition. Hum Reprod 25: 900-907.
- Verkauf BS (1987) Incidence, symptoms, and signs of endometriosis in fertile and infertile women. J Fla Med Assoc 74: 671-675.
- Brewer CJ, Balen AH (2010) The adverse effects of obesity on conception and implantation. Reproduction 140: 347-364.
- Sheh A, Chaturvedi R, Merrellf DS, Correab P, Wilson KT, et al. (2013) Phylogeographic Origin of Helicobacter pylori Determines Host-Adaptive Responses upon Coculture with Gastric Epithelial Cells. Infection and Immunity 81: 2468-2477.
- Delitala AP, Pes GM, Malaty HM, Pisanu G, Dore MP, et al. (2016) Implication of Cytotoxic Helicobacter pylori Infection in Autoimmune Diabetes. Journal of Diabetes Research.
- Kostic AD, Xavier RJ, Gevers D (2014) The Microbiome in Inflammatory Bowel Disease: Current Status and the Future Ahead. Gastroenterology 146: 1489-1499.
- Potgieter M, Bester J, Kell DB, et al. (2015) The dormant blood microbiome in chronic, inflammatory diseases. Fems Microbiology Reviews 39: 567-591.
- Sheehan D, Shanahan F (2017) The Gut Microbiota in Inflammatory Bowel Disease. Gastroenterology Clinics of North America 46: 143.
- Smith BC, Chen Z, Harari A, Barris DM, Viswanathan S, et al. (2012) The Cervical Microbiome over 7 Years and a Comparison of Methodologies for Its Characterization. Plos One 7: e40425.
- Jumpstart Consortium Human Microbiome Project Data Generation Working, G (2012) Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One 7.
- Sipos M, Jeraldo P, Chia N, Ani Qu A, Singh D, et al. (2010) Robust Computational Analysis of rRNA Hypervariable Tag Datasets. Plos One 5: e15220.
- Ye Y (2011) Identification and Quantification of Abundant Species from Pyrosequences of 16S rRNA by Consensus Alignment. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2010: 153-157.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushmanet FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335-336.
- Harwich MD, Serrano MG, Fettweis JM, Alves João MP, Reimerset MA, et al. (2012) Genomic sequence analysis and characterization of Sneathia amnii sp nov. Bmc Genomics 13: S4.
- Payne MS and Bayatibojakhi S (2014) Exploring preterm birth as a polymicrobial disease: an overview of the uterine microbiome. Frontiers in Immunology 5: 1-12.
- (1997) Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril 67: 817-821.
- Hyman RW, Bernstein D, Vo KC, Zelenko Z, Davis RW, et al. (2012) The dynamics of the vaginal microbiome during infertility therapy with in vitro fertilization-embryo transfer. J Assist Reprod Genet 29: 105-115.
- Harirchian P, Gashaw I, Lipskind ST, Braundmeier AG, Hastings JM, et al. (2012) Lesion kinetics in a non-human primate model of endometriosis. Hum Reprod 27: 2341-2351.
- Braundmeier AJ, Hastings JM, Fazleabas AT (2008) Endometriosis alters the peripheral expression of regulatory T cells in a hon-human primate. in American Society for Reproductive Medicine, San Francisco, CA.
- Braundmeier AG, Fazleabas AT (2009) The non-human primate model of endometriosis: research and implications for fecundity. Mol Hum Reprod 15: 577-586.
- Cunningham Hey AJ, Fazleabas AT, Braundmeier AG, et al. (2011) Endometrial stromal cells and immune cell populations within lymph nodes in a nonhuman primate model of endometriosis. Reprod Sci 18: 747-754.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences